Biography
I am a PhD student in Biostatistics at New York University under the supervision of Michele Santacaterrina and Ivan Diaz.
My research focuses methodological development of novel causal inference methods. My primary interests center around developing doubly-robust estimator theory for transportability and generalization of treatment specific survival curves, and developing doubly-robust estimators in the context of repeated continuous/factorial exposures effects.
Prior to joining NYU for a doctoral program I was a research biostatistician at Memorial Sloan Kettering cancer center (MSKCC) in New-York city in the department of Epidemiology & Biostatistics.
During my time there focuses on the development of complex methods for survival prediction and stratification in high-dimensional genomic datasets of various cancer cohorts. Over the course of my time at MSKCC I have developed some open-source R packages to perform ensemble learning for left-truncated genomic survival datasets, including some online visual tools for exploration, validation and individual predictions. This focus on genomics also lead me to develop tools to streamline data retrieval and processing into an analysis ready format, building pipelines enabling beginners to perform complex analytics in a very accessible way.
I also specialize in R Shiny application development both for research and educational purposes in order to render in a more user friendly way complex statistical results.
Download my resumé .
- Causal Inference
- Survival Analysis/Prediction/Stratification with Delayed Entry
- Cancer Genomics
- Optimized Individualized Treatment Rules
- Software development
-
PhD in Biostatistics, Current
New York University
-
MS in Biostatistics, 2017
University of Michigan
-
BS in Mathematics and Computer Science, 2014
McGill University
Projects
Featured Publications
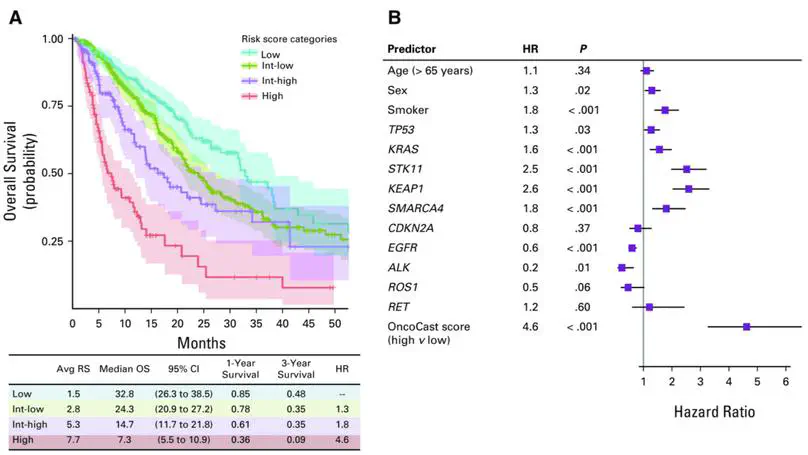
By using sequencing results from a cohort of 1,054 patients with advanced lung adenocarcinomas, we developed OncoCast, a machine learning tool for survival risk stratification and biomarker identification. We found that comutations of both STK11 and KEAP1 are strong determinants of unfavorable prognosis with currently available therapies.
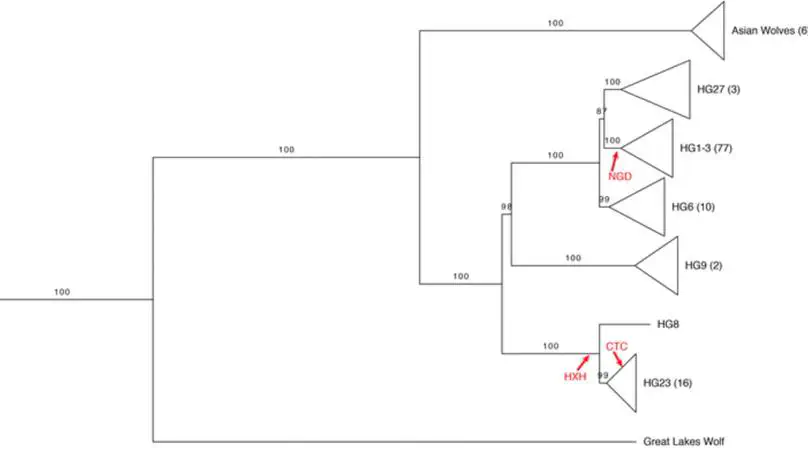
Utilizing a recently published dog Y-chromosome reference, we analyzed Y-chromosome sequence across a diverse collection of canids and determined the Y haplogroup of three ancient European dogs. We estimate that the time to the most recent comment ancestor of dog Y haplogroups is 68–151 thousand years ago and confirm long-standing population structure among European dogs.